 Genomic DNA is a shared substrate for the two principal processes in the cell nucleus – gene transcription and DNA replication. In our lab, we study the fundamental mechanisms of transcription and DNA replication in eukaryotes. We aim to understand what aspects of regulation of transcription and DNA replication differentiate healthy and diseased cells. Ultimately, we hope to reveal specific dependencies thus paving the way for development of new targeted therapies. To achieve these goals, our lab utilizes many approaches including functional genomics, proteomics, biochemistry, and molecular biology. Conservancy of factors and pathways involved in transcription and DNA replication allows us to use the powerful yeast system as our primary model.
Genomic DNA is a shared substrate for the two principal processes in the cell nucleus – gene transcription and DNA replication. In our lab, we study the fundamental mechanisms of transcription and DNA replication in eukaryotes. We aim to understand what aspects of regulation of transcription and DNA replication differentiate healthy and diseased cells. Ultimately, we hope to reveal specific dependencies thus paving the way for development of new targeted therapies. To achieve these goals, our lab utilizes many approaches including functional genomics, proteomics, biochemistry, and molecular biology. Conservancy of factors and pathways involved in transcription and DNA replication allows us to use the powerful yeast system as our primary model.
Recruitment, regulation, and roles of chromatin readers from the BET family
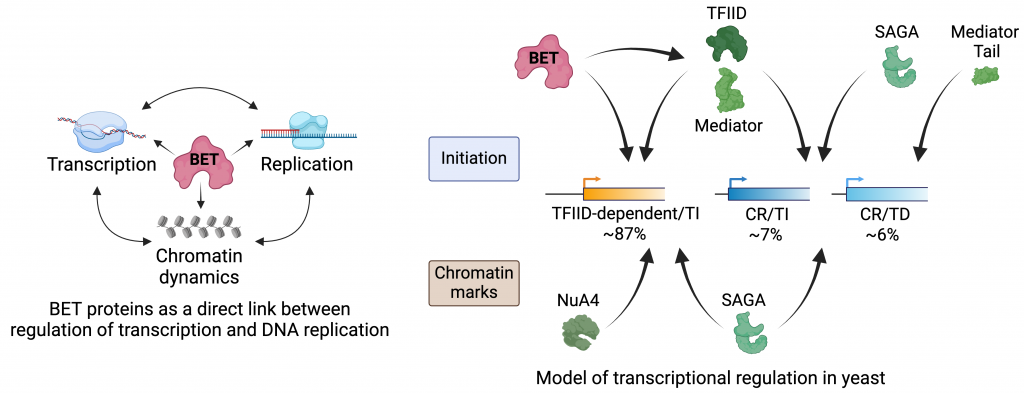
We previously characterized yeast bromodomain-containing factors Bdf1/2 as functional homologues of chromatin readers from the human BET family. Aberrant activity of human BET proteins in transcriptional regulation have been linked with development and progression of many diseases. Canonical models assert that as chromatin readers BET proteins are recruited to regulatory elements by their tandem bromodomains, which ‘read’ acetylated chromatin. However, it is less clear how BET proteins regulate transcription once recruited, and recent evidence suggests that bromodomain activity alone does not fully explain recruitment. For example, diseased cells quickly develop resistance to therapeutics that specifically inhibit bromodomain activity. Further complicating the subject, our recent unpublished findings have implicated BET proteins in regulation of DNA replication. Based on this background, we are now exploring the roles of BET proteins in both transcription and DNA replication. We are investigating the BET protein interaction network, functional crosstalk between BET proteins and coactivators and chromatin modifying complexes, mechanisms of BET protein recruitment to chromatin, specific functions of conserved domains of BET proteins, and the level of a functional redundancy between distinct members of the BET family. The last project involves both yeast and mammalian cells as models.
Spatiotemporal coordination of gene transcription and DNA replication
The maintenance of cellular homeostasis and genome stability depends on the strict control of gene transcription and DNA replication and the spatiotemporal coordination of the two processes to avoid conflicts. A plausible model asserts that transcription and DNA replication are coordinated at the early stages of both processes when respective machineries are assembled and activated. In support of this model, most genes are transcribed during the whole cell cycle, ~50% of origins of replication are located in active gene promoters or promoter-proximal regions, transcriptionally active genes tend to be replicated earlier, and both initiation of transcription and initiation of DNA replication are positively associated with the same chromatin characteristics. Based on this background and our unpublished findings, we are testing a model where BET proteins function as a direct link between genome-wide regulation of transcription and DNA replication. Our preliminary data indicate that BET proteins may facilitate the crosstalk between transcription and DNA replication at least partially through recruitment of distinct protein complexes involved in chromatin dynamics. We are also testing if replication initiation factors directly contribute to the regulation of transcription when cells prepare for DNA replication.